Resistor-capacitor-inductor blood vessel with optional stenosis. More...
#include <BloodVessel.h>
Public Types | |
| enum | ParamId { RESISTANCE = 0 , CAPACITANCE = 1 , INDUCTANCE = 2 , STENOSIS_COEFFICIENT = 3 } |
| Local IDs of the parameters. More... | |
Public Member Functions | |
| BloodVessel (int id, Model *model) | |
| Construct a new BloodVessel object. | |
| void | setup_dofs (DOFHandler &dofhandler) |
| Set up the degrees of freedom (DOF) of the block. | |
| void | update_constant (SparseSystem &system, std::vector< double > ¶meters) |
| Update the constant contributions of the element in a sparse system. | |
| void | update_solution (SparseSystem &system, std::vector< double > ¶meters, const Eigen::Matrix< double, Eigen::Dynamic, 1 > &y, const Eigen::Matrix< double, Eigen::Dynamic, 1 > &dy) |
| Update the solution-dependent contributions of the element in a sparse system. | |
| void | update_gradient (Eigen::SparseMatrix< double > &jacobian, Eigen::Matrix< double, Eigen::Dynamic, 1 > &residual, Eigen::Matrix< double, Eigen::Dynamic, 1 > &alpha, std::vector< double > &y, std::vector< double > &dy) |
| Set the gradient of the block contributions with respect to the parameters. | |
 Public Member Functions inherited from Block Public Member Functions inherited from Block | |
| Block (int id, Model *model, BlockType block_type, BlockClass block_class, std::vector< std::pair< std::string, InputParameter > > input_params) | |
| Construct a new Block object. | |
| ~Block () | |
| Destroy the Block object. | |
| Block (const Block &)=delete | |
| Copy the Block object. | |
| std::string | get_name () |
| Get the name of the block. | |
| void | update_vessel_type (VesselType type) |
| Update vessel type of the block. | |
| void | setup_params_ (const std::vector< int > ¶m_ids) |
| Setup parameter IDs for the block. | |
| void | setup_dofs_ (DOFHandler &dofhandler, int num_equations, const std::list< std::string > &internal_var_names) |
| Set up the degrees of freedom (DOF) of the block. | |
| virtual void | setup_model_dependent_params () |
| Setup parameters that depend on the model. | |
| virtual void | setup_initial_state_dependent_params (State initial_state, std::vector< double > ¶meters) |
| Setup parameters that depend on the initial state. | |
| virtual void | update_time (SparseSystem &system, std::vector< double > ¶meters) |
| Update the time-dependent contributions of the element in a sparse system. | |
| virtual void | post_solve (Eigen::Matrix< double, Eigen::Dynamic, 1 > &y) |
| Modify the solution after solving it. | |
| virtual TripletsContributions | get_num_triplets () |
| Get number of triplets of element. | |
| virtual void | set_activation_function (std::unique_ptr< ActivationFunction > af) |
| Set activation function (for chamber blocks that use one). | |
Public Attributes | |
| TripletsContributions | num_triplets {5, 3, 2} |
| Number of triplets of element. | |
 Public Attributes inherited from Block Public Attributes inherited from Block | |
| const int | id |
| Global ID of the block. | |
| const Model * | model |
| The model to which the block belongs. | |
| const BlockType | block_type |
| Type of this block. | |
| const BlockClass | block_class |
| Class of this block. | |
| VesselType | vessel_type = VesselType::neither |
| Vessel type of this block. | |
| const std::vector< std::pair< std::string, InputParameter > > | input_params |
| Map from name to input parameter. | |
| std::vector< Node * > | inlet_nodes |
| Inlet nodes. | |
| std::vector< Node * > | outlet_nodes |
| Outlet nodes. | |
| bool | steady = false |
| Toggle steady behavior. | |
| bool | input_params_list = false |
| Are input parameters given as a list? | |
| std::vector< int > | global_param_ids |
| Global IDs for the block parameters. | |
| std::vector< int > | global_var_ids |
| Global variable indices of the local element contributions. | |
| std::vector< int > | global_eqn_ids |
| Global equation indices of the local element contributions. | |
| TripletsContributions | num_triplets |
| Number of triplets of element. | |
Detailed Description
Resistor-capacitor-inductor blood vessel with optional stenosis.
Models the mechanical behavior of a bloodvessel with optional stenosis.
![\[\begin{circuitikz} \draw
node[left] {$Q_{in}$} [-latex] (0,0) -- (0.8,0);
\draw (1,0) node[anchor=south]{$P_{in}$}
to [R, l=$R$, *-] (3,0)
to [R, l=$S$, -] (5,0)
(5,0) to [L, l=$L$, -*] (7,0)
node[anchor=south]{$P_{out}$}
(5,0) to [C, l=$C$, -] (5,-1.5)
node[ground]{};
\draw [-latex] (7.2,0) -- (8,0) node[right] {$Q_{out}$};
\end{circuitikz}
\]](form_14.png)
Governing equations
![\[P_\text{in}-P_\text{out} - (R + S|Q_\text{in}|) Q_\text{in}-L
\dot{Q}_\text{out}=0 \]](form_15.png)
![\[Q_\text{in}-Q_\text{out} - C \dot{P}_\text{in}+C(R +
2S|Q_\text{in}|) \dot{Q}_{in}=0 \]](form_16.png)
Local contributions
![\[\mathbf{y}^{e}=\left[\begin{array}{llll}P_{i n} & Q_{in} &
P_{out} & Q_{out}\end{array}\right]^\text{T} \]](form_17.png)
![\[\mathbf{F}^{e}=\left[\begin{array}{cccc}
1 & -R & -1 & 0 \\
0 & 1 & 0 & -1
\end{array}\right]
\]](form_18.png)
![\[\mathbf{E}^{e}=\left[\begin{array}{cccc}
0 & 0 & 0 & -L \\
-C & CR & 0 & 0
\end{array}\right]
\]](form_19.png)
![\[\mathbf{c}^{e} = S|Q_\text{in}|
\left[\begin{array}{c}
-Q_\text{in} \\
2C\dot{Q}_\text{in}
\end{array}\right]
\]](form_20.png)
![\[\left(\frac{\partial\mathbf{c}}{\partial\mathbf{y}}\right)^{e} =
S \text{sgn} (Q_\text{in})
\left[\begin{array}{cccc}
0 & -2Q_\text{in} & 0 & 0 \\
0 & 2C\dot{Q}_\text{in} & 0 & 0
\end{array}\right]
\]](form_21.png)
![\[\left(\frac{\partial\mathbf{c}}{\partial\dot{\mathbf{y}}}\right)^{e} =
S|Q_\text{in}|
\left[\begin{array}{cccc}
0 & 0 & 0 & 0 \\
0 & 2C & 0 & 0
\end{array}\right]
\]](form_22.png)
with the stenosis resistance 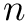
Gradient
Gradient of the equations with respect to the parameters:
![\[\mathbf{J}^{e} = \left[\begin{array}{cccc}
-y_2 & 0 & -\dot{y}_4 & -|y_2|y_2 \\
C\dot{y}_2 & (-\dot{y}_1+(R+2S|Q_\text{in}|)\dot{y}_2) & 0 & 2C|y_2|\dot{y}_2
\end{array}\right]
\]](form_26.png)
Parameters
Parameter sequence for constructing this block
0Poiseuille resistance1Capacitance2Inductance3Stenosis coefficient
Usage in json configuration file
"vessels": [
{
"vessel_id": 0,
"vessel_length": 10.0,
"vessel_name": "branch0_seg0",
"zero_d_element_type": "BloodVessel",
"zero_d_element_values": {
"R_poiseuille": 100.0,
"C": 1.0e-5,
"L": 1.0e-6,
"stenosis_coefficient": 0.0
}
}
]
Internal variables
This block has no internal variables.
Member Enumeration Documentation
◆ ParamId
| enum BloodVessel::ParamId |
Local IDs of the parameters.
Constructor & Destructor Documentation
◆ BloodVessel()
|
inline |
Construct a new BloodVessel object.
- Parameters
-
id Global ID of the block model The model to which the block belongs
Member Function Documentation
◆ setup_dofs()
|
virtual |
Set up the degrees of freedom (DOF) of the block.
Set global_var_ids and global_eqn_ids of the element based on the number of equations and the number of internal variables of the element.
- Parameters
-
dofhandler Degree-of-freedom handler to register variables and equations at
Reimplemented from Block.
◆ update_constant()
|
virtual |
Update the constant contributions of the element in a sparse system.
- Parameters
-
system System to update contributions at parameters Parameters of the model
Reimplemented from Block.
◆ update_gradient()
|
virtual |
Set the gradient of the block contributions with respect to the parameters.
- Parameters
-
jacobian Jacobian with respect to the parameters alpha Current parameter vector residual Residual with respect to the parameters y Current solution dy Time-derivative of the current solution
Reimplemented from Block.
◆ update_solution()
|
virtual |
Update the solution-dependent contributions of the element in a sparse system.
- Parameters
-
system System to update contributions at parameters Parameters of the model y Current solution dy Current derivate of the solution
Reimplemented from Block.
Member Data Documentation
◆ num_triplets
| TripletsContributions BloodVessel::num_triplets {5, 3, 2} |
Number of triplets of element.
Number of triplets that the element contributes to the global system (relevant for sparse memory reservation)
The documentation for this class was generated from the following files:
- model/BloodVessel.h
- model/BloodVessel.cpp
Generated by